Antimicrobials or antibiotics were the important revelations of the last century, however, it came along with a silent curse that people care less to talk about. Antimicrobial resistance (AMR) which emerged alongside antibiotics in the last century has been a significant concern for scientists and policymakers. Since their discovery, it has been noted that the widespread use of antibiotics is the primary cause of bacteria developing antimicrobial drug resistance. Despite the recognition of this issue, it is challenging to curtail the widespread use of antibiotics because they are essential for treating various infections. Paradoxically, the necessity of using these drugs becomes an inadvertent advantage for bacteria to evolve resistance mechanisms. This dilemma creates a seeming stalemate in our battle against these tiny microorganisms. Delaying action could have dire consequences, potentially leading to the emergence of stronger superbugs that pose a serious threat to the entire human population. The recent COVID-19 pandemic serves as a stark reminder of the devastating impact a small microbe can have on global health. This paper delves into the mechanisms of antimicrobial resistance in bacteria, the evolution of superbugs and the innovative techniques employed by scientists to combat these challenges. Taking proactive steps is crucial to avoid a future where we are at the mercy of increasingly resilient microbes.
Antimicrobial Resistance, Superbugs, Efflux pumps, β-Lactamases, Nanoparticles, CRISPR/Cas9
Antibiotics were systematically thought to be the important discovery of the previous century. Their boundless use transformed healthcare after their debut and the names of scientists like Paul Ehrlich and Sir Alexander Fleming1 who introduced them to us are still discussed. Various antibiotics have effectively controlled infectious diseases that were once untreatable and fatal, thus preventing millions of deaths each year.2 However, due to the overuse and misuse of antibiotics, the effectiveness of these drugs is now threatened by the rise of antibiotic resistant bacteria. Fleming, in his days, had noted the insensitive use of these drugs might result in the development of bacterial strains which might become immune.3 This ability of microorganisms, such as bacteria, viruses and some parasites, to develop resistance to antimicrobial drugs has been termed antimicrobial resistance (AMR).4 This resistance means that the antimicrobial drugs, including antibiotics, antivirals and antiparasitic medications, are no longer as effective or entirely ineffective in killing or inhibiting the growth of these microorganisms. As a result, infections become more challenging to treat, leading to prolonged illnesses, increased healthcare costs and in some cases, higher mortality rates.5
The World Health Organization (WHO) has recognized AMR as one of the ten major worldwide public health challenges.6 It is a significant global concern that requires coordinated efforts to combat its spread and ensure the continued effectiveness of these vital drugs. This has prompted researchers and pharmaceutical companies to intensify their efforts in drug research and development, with a focus on understanding resistance mechanisms and developing novel antibiotics and alternative treatment strategies. In this article, we have explored the various AMR mechanisms and discussed methods to prevent it.
Antimicrobials
Antimicrobials are a diverse group of substances that are pivotal in the fight against infectious diseases.7 These compounds, including antibiotics, antivirals, antifungals and antiparasitic, work by either killing or inhibiting the growth of microorganisms such as bacteria, viruses, fungi and parasites.8 They play a crucial role in medicine, agriculture and veterinary care, combating infections in humans, animals and plants. Antimicrobials encompass a wide array of compounds produced through various means, including natural sources like bacteria, fungi and plants, as well as synthetic processes in laboratories.9 Their mechanisms of action vary depending on the specific antimicrobial and the type of microorganism it targets. For example, bactericidal antimicrobials, such as penicillin, cephalosporins and vancomycin directly kill the bacteria. However, bacteriostatic antimicrobials on the other hand, such as chloramphenicol, tetracyclines and erythromycin, inhibit the growth of bacterial cells
AMR
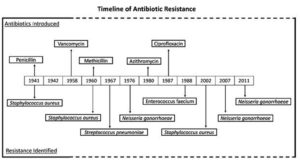
The initial indication of AMR became evident shortly following the identification of penicillin.10 In 1940, Abraham and Chain observed that a strain of E. coli had the capacity to render penicillin ineffective through the production of penicillinase.11 This early discovery laid the foundation for our understanding of resistance mechanisms. The timeline of antibiotic resistance development for different drugs are mentioned in Figure 1. When penicillin and sulphonamide were introduced in medical treatments during the late 30s and 40s of the 20th centuries, individuals believed that antibiotic use prevented every disease that is caused by the disease-causing agents. But the exploitation of these antibiotics has caused tremendous pressure in the development and improvement of AMR.12 Different bacteria started to develop resistance mechanism against the antibiotics and a few bacteria along with their mechanism of actions are mentioned in Table 1. The momentum for improving antibiotic resistance has been growing, especially as the use of antibiotics became more restricted after vancomycin was reserved for special cases, preventing resistance from developing. However, irregular antibiotic usage brought about the rise of methicillin-resistant Staphylococcus aureus (MRSA), which shows multi antimicrobial resistance. Consequently, vancomycin-resistant Enterococci (VRE) showed up in 1986 because of the far-reaching utilization of vancomycin, which was viewed as the antimicrobial after all other options had run out. AMR is primarily caused by the overuse and misuse of antimicrobial drugs. Over-prescription of antibiotics for viral infections, self-medication and over-the-counter sales of antibiotics contribute to the problem.13 In agriculture, antibiotics are not only used to treat sick animals but are also commonly added to the feed and drinking water of healthy animals to prevent illness.14 There is substantial evidence indicating that the use of antibiotics in animals significantly contributes to the development of AMR in human pathogens. Additionally, the slow development of new antimicrobial drugs, coupled with poor infection control practices in healthcare settings and communities, allows drug resistant pathogens to thrive and spread.15 These factors collectively contribute to the emergence and proliferation of AMR, making it a pressing global health concern. To battle these hazardous issues, it is essential for the continual efforts to develop new antimicrobial agents. Some recent antibiotics have been proven to be powerful upon MRSA and VRE, but these dangerous microorganisms will eventually develop resistance to these compounds.
Table (1):
Overview of Antibiotic Resistance Mechanism Employed by Bacteria
Antibiotic |
Target |
Mechanism Of Action |
Mechanism Of Resistance |
Ref. |
|---|---|---|---|---|
β-Lactams |
Penicillin-binding proteins (PBPs) |
Inhibits bacterial cell wall synthesis by binding to PBPs, preventing cross-linking of peptidoglycan chains. |
Production of β-lactamases that hydrolyse the β-lactam ring. Alteration or decrease in expression of PBPs. |
59 |
Vancomycin |
D-Ala-D-Ala terminus of peptidoglycan precursors |
Sequestration of substrate required for cross linking and cell wall synthesis. |
Reprogramming of D-Ala-D-Ala to D-Ala-D-Lac or D-Ala –D-ser. |
60 |
Macrolides (Erythromycin Class) |
Bacterial ribosomes (50S subunit) |
Inhibits bacterial protein synthesis by binding to 50S ribosomal subunit, preventing tRNA translocation. |
Modification of target site through methylation. Active efflux of antibiotic from the cell. |
61 |
Tetracyclines |
Bacterial ribosomes (30S subunit) |
Inhibits bacterial protein synthesis by binding to 30S ribosomal subunit, preventing aminoacyl-tRNA binding. |
Active efflux of antibiotic. Production of proteins that protect ribosomal binding sites. |
62 |
Aminoglycosides |
Bacterial ribosomes (30S subunit) |
Inhibits bacterial protein synthesis by causing misreading of mRNA and incorporation of incorrect amino acids. |
Modification of antibiotic through acetylation, phosphorylation, or adenylation. Decreased uptake or increased efflux of antibiotic. |
63 |
Oxazolidinones |
Bacterial ribosomes (50S subunit) |
Inhibits bacterial protein synthesis by preventing the formation of the initiation complex. |
Mutations in the 23S rRNA of the ribosome, reducing the binding affinity of antibiotic. |
64 |
Fluoroquinolones |
DNA gyrase and topoisomerase IV |
Inhibits bacterial DNA synthesis and replication by interfering with DNA gyrase and topoisomerase IV. |
Mutations in target enzymes Active efflux of antibiotic from the cell. |
65 |
AMR mechanisms
The resistance to antimicrobials is either inherited or intrinsic. For instance, in the natural resistance mechanism caused by Pseudomonas aeruginosa, the lower membrane permeability would be the probable reason for its intrinsic protection from numerous antimicrobials. Inherited resistance happens by securing a transposon or a plasmid that carries the genes that encode resistance mechanisms from other organisms, or via the process of chromosomal transformation.
The significant mechanisms of actions involved in the development of microbial resistance are:
- Reduction in drug binding affinity by altering drug targets.
- Modification of antibiotics causing inactivation of drugs.
- Increasing efflux or restricting uptake.
- Genetic mutations.
Modification of drug targets
The modification of drug targets is a crucial mechanism that microorganisms employ to develop AMR. This mechanism involves altering certain segments or structures in the microorganism that are the targets of the antimicrobial agents. For example, the change in the number and structure of penicillin-binding proteins (PBPs) is a major component of protection that is utilized by gram-negative microbes.16
PBPs are enzymes found in bacterial cell walls that play a key role in the synthesis and maintenance of the cell wall structure. Antibiotics like penicillin exert their antimicrobial effects by binding to these PBPs, thereby inhibiting cell wall synthesis and leading to bacterial cell death. However, when there is an increase in the production of PBPs or alterations in their structure, the binding of penicillin to these proteins may be hindered, reducing the drug’s effectiveness.9 Another example of drug target modification seen in gram-negative bacteria, where resistance to vancomycin occurs due to changes in the structure of peptidoglycan precursors.17 Other mechanism that have been reported include alterations in the surface charge of the bacterial cell membrane, such as a shift towards a positive charge which can interfere with the binding of calcium ions, thereby reducing the efficacy of certain drugs such as daptomycin.18
Inactivation of drugs
The two fundamental manners by which microorganisms stop the activity of drugs are by either transferring a chemical group to the drug or by directly degrading the drug.19 One of the most well-known examples of drug inactivation is the production of β-lactamase enzymes by many bacteria. β-lactamases are an exceptionally enormous category of drug hydrolysing enzymes. These enzymes target and inactivate β-lactam antibiotics, which include widely used drugs like penicillin and cephalosporins. β-lactam antibiotics work by interfering with the synthesis of the bacterial cell wall, ultimately causing the cell to burst. However, β-lactamase enzymes can break the β-lactam ring, a key structural component of these antibiotics, rendering them inactive.20 As a result, the antibiotic can no longer disrupt the cell wall synthesis and the bacterium remains unharmed.
Tetracycline is another drug that can be easily hydrolysed and inactivated by the tetX enzyme which modifies the tetracycline molecule by adding a functional group to specific sites on the tetracycline structure. This change in structure interferes with the tetracycline’s ability to bind to the ribosome effectively. As tetracycline antibiotics depend on their specific interaction with the ribosomes to inhibit protein synthesis, the altered tetracycline molecule can no longer bind to the ribosome with the same affinity or effectiveness.21 As a result, it cannot disrupt the translation process and bacterial protein synthesis proceeds unimpeded.
The inactivation of certain drugs is brought about by a group of transferases responsible for transferring chemical groups such as phosphoryl, adenyl and acetyl to these drugs.9 The most utilized mechanism is acetylation, which shows its effect against drugs like streptogramins, aminoglycosides and fluoroquinolones.
Active efflux
Many microorganisms have evolved with an amazing mechanism to counter increasing drug concentration levels in their cells.22 They simply efflux the antibacterial agents outside the cells through certain pumps in their cell surface known as efflux pumps.23 Via these molecular pumps, they can actively transport a wide variety of antimicrobial compounds and toxins out of the cells.24 Efflux pumps can exhibit specificity towards a single substrate or have the ability to transport a variety of structurally dissimilar compounds. This includes antibiotics from various classes, and these pumps may be linked to the phenomenon of multiple drug resistance (MDR).25
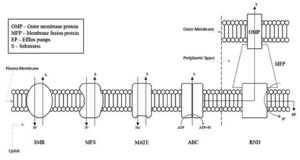
The efflux pumps come under five families which are:
- ATP-Binding Cassette (ABC) superfamily
- Multidrug and Toxic Compound Extrusion (MATE) superfamily
- Small Multidrug Resistance (SMR) superfamily
- Major Facilitator Superfamily (MFS)
- Resistance Nodulation and Cell Division (RND) superfamily
One of the major differences between the different families is the varying energy sources and assembly of the components as outlined in Figure 2. The ABC family relies on ATP hydrolysis for energy, as opposed to other transporters like MATE, MFS, RND and SMR superfamilies, which harness the proton-motive force from H+ or the electrochemical gradient of Na+ for energy to expel various compounds.26 Another difference as seen in the RND family is the tripartite complex comprising of the outer-membrane canal protein (OMP), an inner-membrane transporter and a membrane fusion protein (MFP) that bridges the OMP and inner-membrane transporter.26
The genes are chromosomally encoded and some of these are constitutively expressed while others are overexpressed under certain conditions like when the availability of a substrate is reasonable.27
Genes associated with efflux pumps can be obtained via intrinsic or acquired means. Certain bacteria possess these genes on their chromosomes, providing an inherent survival mechanism in challenging environments.28 In contrast, other bacteria can procure these genes through diverse mechanisms such as mutations within local repressor genes, activation of a regulon controlled by a global transcriptional regulator, or the presence of efflux pump genes on plasmids.28 For example, the RND family efflux pumps play a crucial role in the intrinsic antibiotic resistance of gram-negative bacteria by actively expelling a diverse array of antibiotics and antimicrobials.29
In gram-positive bacteria, the efflux pumps provide intrinsic resistance as they are encoded on the chromosome.23 Chromosomal encoding is responsible for MDR efflux pumps, exemplified by NorA, NorB, MepA and MdeA in S. aureus.30 These pumps confer inherent antibiotic resistance in bacteria across a diverse spectrum. Conversely, certain efflux pumps in gram-positive are also carried around on plasmids or transposons, like QacA/B in S. aureus or MefA and MefB in Streptococcus spp., respectively.30
Genetic mutations
In specific species, mutation is the primary, or sole, reason for AMR. Perhaps the best example would be Mycobacterium tuberculosis, where mutation is the primary cause of resistance to all clinical drugs in this bacterium. Combination treatment is required to treat tuberculosis which is a pathogen that causes prolonged sickness in humans.31 The risk caused by the mutation at the point of infection in microorganisms cannot be eliminated by the combination treatment which only diminishes the impact.
Alteration in their genome will prevent the ability of antibiotics to act on them thus creating a superbug or super bacteria. These naturally genetically modified bacteria produce resistance against the exploited drugs which then spread among their population leading to the entire species becoming resistant to certain drugs.19 Moreover, in some cases, after a bacterium dies, they may leave their genetic material outside which in turn might be picked up by bacteria of a different species that might incorporate the usable resistant genetic material inside it resulting in producing new resistance.
Superbugs
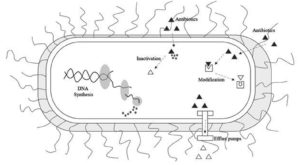
Superbugs signify strains of MDR bacteria that are impervious to a large portion of the antibiotics and different drugs usually used to treat the disease they cause.32 The expression “superbug” was coined by the media and essentially refers to bacteria that has developed resistance to multiple drugs that were once effective against the infection it caused.33 Different methods of resistance mechanism developed by bacteria are mentioned in Figure 3. A couple of instances of superbugs include MRSA, VRE, multidrug-resistant Pseudomonas aeruginosa and multidrug-resistant Acinetobacter.34 It is crucial to understand that any bacteria have the potential to evolve into a superbug and develop resistance to nearly any drug if those drugs are overused or misused. This misuse can have harmful consequences for crops, humans and animals. Thus leading to a situation where a multi-step approach may be the only effective approach to further prevent the spread and infection of these superbugs.35
Development of superbugs
The development of superbugs is primarily due to the misuse and overuse of antibiotics, which leads to the emergence of drug resistant strains.36 When antibiotics are overused or misused, the microorganisms that cause the infections tend to develop resistance to multiple antibiotics via the various mechanisms that have been discussed earlier in this article. As the microorganisms evolve to survive the drugs that were previously lethal, they create antibiotic resistant strains and commonly used antibiotics become ineffective over time.36
The emergence and spread of superbugs result from a combination of factors such as the overuse and misuse of antibiotics, inappropriate antibiotic prescribing practices and incomplete treatment courses.37 Nowadays the most common form of antibiotic misuse is consumption of antibiotics without consulting physicians. Furthermore, there are instances where healthcare professionals prescribe antibiotics unnecessarily. This may result from diagnostic uncertainty, patient requests, or precautionary practices. In cases where antibiotics are appropriately prescribed, some patients may not complete the prescribed course.38 Incomplete courses can lead to the survival of bacteria that develop resistance to the antibiotic. The reason for this lies in the fact that the most susceptible bacteria are eliminated first, leaving behind those that may possess greater resistance. Additionally, improper and excessive use of antimicrobials in livestock, fish farming and agriculture leads to the development of resistant bacteria, which causes major health risks.39,40
Combating AMR
AMR may be a complicated issue with various causative factors. It’s the major reason behind health issues in the community, affecting people directly or indirectly. Given the necessity for widespread antibiotic use to combat AMR, the immediate development of new technology is imperative.
The overuse of certain antibiotics may result in the production of even more powerful superbugs which might ultimately become resistant to all known drugs.31 Hence new techniques to combat AMR is needed and to our surprise, many alternative techniques to combat AMRs have been developed by scientist all around the world41 to fight against the globally spreading AMR.
Targeting outer membrane
Bacteria have evolved various strategies to avoid contact with antibiotics, and one crucial mechanism involves preventing the entry of these drugs.9 Antibiotics typically exert their effects after entering the cell membrane, but bacteria have developed defenses to impede this process.41 The intricate structure of bacterial cell walls, comprising compounds like porins and lipopolysaccharides, serves as a barrier, hindering the entry of certain antibiotics. These compounds act as gatekeepers, preventing the intrusion of antibiotics.42 Few microbes undergo macromolecule mutations outside the cell membranes to prevent the entry of some antibiotics. However, certain antibiotics have been devised with means to overcome these barriers and enter the organisms.
Hydrophobic compounds
Certain compounds such as macrolides and rifampicin possess hydrophobic characteristics that allows the compounds to navigate through the cell membrane efficiently and easily cross the lipid bilayer.
Hydrophilic molecules
Antibiotics such as β-lactams, fluoroquinolones and phenicol antibiotics, are hydrophilic in nature. They leverage transport mechanisms by interacting with specific porins, allowing them to diffuse through the cell membrane and overcome the protective barriers.
Targeting β–Lactamases
Synthesis of enzymes that can counter the antibiotics administered is one of the major defense mechanisms employed by bacteria.1 β-lactam antibiotics which possess natural bactericidal properties were initially developed and widely used. However, the extensive use of β-lactam antibiotics 9 led to the rapid development of resistant strains in gram-positive bacteria. The primary mechanism of β-lactam antibiotics involves the inactivation of transpeptidases, disrupting the final step in membrane biogenesis.31 Subsequently, bacteria developed mechanisms to evade β-lactam treatment such as reduced expression of transport proteins, increase of efflux pumps and expression of β-lactamases.43 As β-lactamases are enzymes that hydrolyse and disrupt β-lactams, this results in the inactivation of antibiotics that have the functional β-lactam group.31
Several strategies can be employed to address this challenge. Firstly, developing inhibitors specifically designed to target and neutralize β-lactamases can help in preventing the degradation of β-lactam antibiotics. Additionally, a promising strategy is the use of combination therapy, wherein β-lactam antibiotics are administered alongside these newly developed inhibitors, enhancing the antibiotics’ effectiveness.44 Structural modifications of antibiotics represent another avenue, focusing on engineering changes that make these drugs less susceptible to β-lactamase activity. An example would be a β-lactamase-activated antibacterial prodrug where the prodrug only exhibits bactericidal properties upon activation by β-lactamase.45 By employing these methods, antibiotic resistance can be leveraged to specifically target bacteria that produce β-lactamase.
Targeting efflux pumps
The development of efflux pump inhibitors is one of the widely used strategies to combat resistance through efflux.46 These inhibitors are tiny molecules that attach themselves to the pumps and block the expulsion of the antibiotic compounds from the cell.27 An overview of several antibiotic efflux pump families, the specific antibodies associated with the efflux pumps and their inhibitors are mentioned in Table 2. As these small molecules do not possess intrinsic bactericidal activities, they have been tested for synergistic effects at various concentrations in combination with antibiotics.22
Many ways have been explored for inhibiting efflux pumps, including47:
- Disrupting channel protein assembly.
- Interfering with efflux pump gene expression.
- Preventing recognition by adding functional chains.
- Developing small molecules as substrates to block activity of efflux pumps.
Therefore, inhibition of efflux may result in several positive outcomes, including47:
- Maintaining drug concentrations at therapeutic doses.
- Reducing multi-drug resistance.
- Reducing treatment periods.
- Enhancing the activity of antibiotics susceptible to efflux.
Table (2):
Antibiotic Efflux Pump Families, their Substrates and Inhibitors
Efflux Pump Family |
Substrates |
Efflux Pump Inhibitors |
Ref. |
|---|---|---|---|
ATP-Binding Cassette (ABC) superfamily |
Tetracyclines Fluoroquinolones Macrolides Rifampicin Chloramphenicol |
Reserpine |
66, 67 |
Multidrug and Toxic Compound Extrusion (MATE) superfamily |
Fluoroquinolones Tigecycline Pentamidine |
Verapamil |
67, 68 |
Small Multidrug Resistance (SMR) superfamily |
Tetracyclines Erythromycin Sulfadiazine |
66, 67 |
|
Major Facilitator Superfamily (MFS) |
Tetracyclines Fluoroquinolones Erythromycin Rifampicin Chloramphenicol |
Phenothiazines Thioridazines Chalcones Piperine Flavonoid’s Quercetin |
66, 67 |
Resistance Nodulation and Cell Division (RND) superfamily |
Tetracyclines Fluoroquinolones Erythromycin Rifampicin β-Lactams Chloramphenicol |
Phenyl-arginine-β-napthylamide (PaβN) Hydantoins |
66, 67 |
Novel approaches to combat AMR
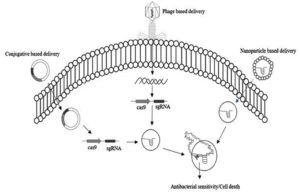
Antibiotic resistance poses a significant threat to global human health.8 Imperative measures are needed to prevent the emergence and spread of multi-drug resistance organisms which could have far-reaching consequences.41 Novel methods to identify, validate and develop new techniques are necessary to overcome this serious issue.16 The latest breakthrough in addressing AMR is using genetic engineering tools. These tools, such as CRISPR/Cas9 technology and nanotechnology, can be used to directly target the antibiotic resistance genes of microorganisms. Figure 4 illustrates various delivery mechanisms for these techniques, which are currently at the forefront of efforts to combat antibiotic resistance.
CRISPR/Cas9 to combat AMR
CRISPR-Cas systems are known as bacterial adaptive systems and are analogues of the immune system.48 The CRISPR/Cas9 system has been employed in many fields of study, ranging from genome editing for agricultural purposes to disease eradication. Originally a natural defense mechanism employed by the bacteria to fight invading phage viruses, scientists have harnessed and replicated these techniques to develop a technology aimed at preventing bacterial impact on humans.49 Bacterial genes contain CRISPR/Cas arrays, which consist of repeating sequences of DNA, known as repeats which are separated by unique sequences of equal sizes known as spacers.50 In the context of combating AMR, CRISPR/Cas9 technique is specifically programmed to target the DNA at specific sites, consequently cleaving AMR genes resulting in the disruption of the bacterial cells’ AMR mechanism.51
Nanoparticles as a tool to combat AMR
Nanotechnology is an emerging field that utilizes nanoparticles to perform multiple or specific functions.52 These nanoparticles can be customized to perform specific tasks. In the context of AMR, utilizing nanoparticles offers a promising approach and unique advantages to address challenges faced by traditional antimicrobials.53 Nanomaterials have the capability to interfere with bacterial membranes, target intracellular components, and facilitate the delivery of antimicrobial agents.54 This results in improved therapeutic effectiveness against stubborn MDR infections.55 The use of nanotechnology alongside CRISPR/Cas9 holds promise for advancing treatments against MDR bacterial infections.56 As mentioned earlier, CRISPR/Cas9, known for its precise targeting, can disrupt drug resistance genes used by microbes for infection or directly eliminate pathogens.57 However, despite its efficacy, challenges in in-vivo delivery efficiency have limited its broader application. Nanotechnology provides a solution by enhancing the effectiveness and safety of CRISPR/Cas9 components through specialized nanoparticle delivery systems, addressing these delivery shortcomings.58
AMR poses a severe global threat that could lead to a catastrophe if not promptly addressed. The most effective strategy against disease caused by bacterial pathogens is not by developing antibiotics to eliminate the pathogens but also to employ necessary measures to control the use of antibiotics. It is crucial for individuals worldwide to contribute to the fight against AMR by preventing the misuse of antibiotics in human and animal treatment. Since this cannot be accomplished overnight, this article offers several unique and successful approaches to combat MDR bacteria. Along with that, this paper has covered some important topics, such as the mechanism of antimicrobial drug resistance and the development of superbugs. While the development of new techniques to fight against AMR may incur significant costs that may be prohibitive for ordinary individuals, the emphasis should be on regulating antibiotic usage. Addressing the crisis of AMR involves raising public awareness on antibiotic misuse and overuse. Accessible diagnostic tools play a crucial role in ensuring that the proper antibiotics are prescribed to the right patient at the correct time, in regulated dosages for a particular time frame until their immune system effectively responds to the infection. Implementing antibiotic stewardship programs and regulatory measures are essential to control antibiotic use in both medical and agricultural settings. These programs can aid in ensuring that there is no room for bacteria to develop resistance mechanisms, ultimately preventing a widespread crisis.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made substantial, direct and intellectual contributions to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Gonzalez-Candelas F, Comas IT, Luis Martinez JA, Carlos Galan J, Baquero F. The Evolution of Antibiotic resistance. Genetics and Evolution of Infectious Diseases. 2011;305-337.
Crossref - Flasche S, Atkins KE. Balancing Benefits and Risks of Antibiotic Use. J Infect Dis. 2018;218(9):1351.
Crossref - Rosenblatt-Farrell N. The landscape of antibiotic resistance. Environ Health Perspect. 2009;117(6):A244-50.
Crossref - Aljeldah MM. Antimicrobial Resistance and Its Spread Is a Global Threat. Antibiotics. 2022;11(8):1082.
Crossref - Murray CJL, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629-655.
Crossref - Antimicrobial resistance. Accessed November 6, 2023. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance
- Kresge N, Simoni RD, Hill RL. Selman Waksman: the Father of Antibiotics. J Biol Chem. 2004;279(48):e7-e8.
Crossref - Pieren M, Tigges M. Adjuvant strategies for potentiation of antibiotics to overcome antimicrobial resistance. Curr Opin Pharmacol. 2012;12(5):551-555.
Crossref - Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482-501.
Crossref - Lobanovska M, Pilla G. Penicillin’s Discovery and Antibiotic Resistance: Lessons for the Future? Yale J Biol Med. 2017;90(1):135-145.
- Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. 1940. Rev Infect Dis. 1988;10(4):677-678.
- Xu X, Xu L, Yuan G, Wang Y, Qu Y, Zhou M. Synergistic combination of two antimicrobial agents closing each other’s mutant selection windows to prevent antimicrobial resistance. Sci Rep. 2018;8(1):1-7.
Crossref - Dadgostar P. Antimicrobial Resistance: Implications and Costs. Infect Drug Resist. 2019;12:3903-3910.
Crossref - Hao H, Cheng G, Iqbal Z, et al. Benefits and risks of antimicrobial use in food-producing animals. Front Microbiol. 2014;5:288.
Crossref - Pulingam T, Parumasivam T, Gazzali AM, et al. Antimicrobial resistance: Prevalence, economic burden, mechanisms of resistance and strategies to overcome. Euro J Pharm Sci. 2022;170:106103.
Crossref - Yoneyama H, Katsumata R. Antibiotic resistance in bacteria and its future for novel antibiotic development. Biosci Biotechnol Biochem. 2006;70(5):1060-1075.
Crossref - Cox G, Wright GD. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013;303(6-7):287-292.
Crossref - Stefani S, Campanile F, Santagati M, Mezzatesta ML, Cafiso V, Pacini G. Insights and clinical perspectives of daptomycin resistance in Staphylococcus aureus: A review of the available evidence. Int J Antimicrob Agents. 2015;46(3):278-289.
Crossref - Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol. 2018;9:2928.
Crossref - Bush K, Bradford PA. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb Perspect Med. 2016;6(8):a025247.
Crossref - Markley JL, Wencewicz TA. Tetracycline-Inactivating Enzymes. Front Microbiol. 2018;9:1058.
Crossref - Martinez JL, Mart L, Fajardo A, Alvarez-ortega C. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev. 2009;33(2):430-449.
Crossref - Piddock LJV. Clinically Relevant Chromosomally Encoded Multidrug Resistance Efflux Pumps in Bacteria. Clin Microbiol Rev. 2006;19(2):382-402.
Crossref - Webber MA, Piddock LJ V. The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemother. 2003;51(1):9-11.
Crossref - Du D, Wang-Kan X, Neuberger A. et al. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol, 2018; 16:523-539.
Crossref - Sreekantan AP, Rajan PP, Mini M, Kumar P. Multidrug Efflux Pumps in Bacteria and Efflux Pump Inhibitors. Advancements of Microbiology. 2022;61(3):105-114.
Crossref - Soto SM. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. 2013;14(3):223-229.
Crossref - Garcia IR, de Oliveira Garcia FA, Pereira PS, et al. Microbial resistance: The role of efflux pump superfamilies and their respective substrates. Life Sci. 2022;295:120391.
Crossref - Athar M, Gervasoni S, Catte A, et al. Tripartite efflux pumps of the RND superfamily: what did we learn from computational studies? Microbiology (NY). 2023;169(3).
Crossref - Costa SS, Viveiros M, Amaral L, Couto I. Multidrug Efflux Pumps in Staphylococcus aureus: an Update. Open Microbiol J. 2013;7(1):59-71.
Crossref - Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect. 2007;13(1):5-18.
Crossref - Painuli S, Semwal P, Sharma R, Akash S. Superbugs or multidrug resistant microbes: A new threat to the society. Health Sci Rep. 2023;6(8):e1480.
Crossref - Algammal A, Hetta HF, Mabrok M, Behzadi P. Editorial: Emerging multidrug-resistant bacterial pathogens “superbugs”: A rising public health threat. Front Microbiol. 2023;14:1135614.
Crossref - Kim D, Jeong SH. Current status of multidrug-resistant bacteria. Journal of the Korean Medical Association. 2022;65(8):468-477.
Crossref - Gray DA, Wenzel M. Multitarget Approaches against Multiresistant Superbugs. ACS Infect Dis. 2020;6(6):1346-1365.
Crossref - Mir S, Brett D, Adam de la B, Martha K. Antibiotics Overuse and Bacterial Resistance. Ann Microbiol Res. 2019;3(1).
Crossref - Mohsin S, Amin MN. Superbugs: a constraint to achieving the sustainable development goals. Bull Natl Res Cent. 2023;47(1):63.
Crossref - Delia SC. Misuse of Antibiotics and the Dissemination of Antibiotic Resistant Bacteria in the Community. MCAST Journal of Applied Research & Practice. 2020;4(1):91-106.
Crossref - Hosain MdZ, Kabir SML, Kamal MdM. Antimicrobial uses for livestock production in developing countries. Vet World. 2021;14(1):210-221.
Crossref - Ibrahim M, Ahmad F, Yaqub B, et al. Current trends of antimicrobials used in food animals and aquaculture. Antibiotics and Antimicrobial Resistance Genes in the Environment. 2020:39-69.
Crossref - Bilal M, Rasheed T, Iqbal HMN, Hu H, Wang W, Zhang X. Macromolecular agents with antimicrobial potentialities: A drive to combat antimicrobial resistance. Int J Biol Macromol. 2017;103:554-574.
Crossref - Potts M. Mechanisms of desiccation tolerance in cyanobacteria. Eur J Phycol. 1999;34(4):319-328.
Crossref - Cole MS, Hegde PV, Aldrich CC. β-Lactamase-Mediated Fragmentation: Historical Perspectives and Recent Advances in Diagnostics, Imaging, and Antibacterial Design. ACS Infect Dis. 2022;8(10):1992-2018.
Crossref - Yan Y, Li G, Li G. Principles and current strategies targeting metallo-β-lactamase mediated antibacterial resistance. Med Res Rev. 2020;40(5):1558-1592.
Crossref - Evans LE, Krishna A, Ma Y, et al. Exploitation of Antibiotic Resistance as a Novel Drug Target: Development of a β-Lactamase-Activated Antibacterial Prodrug. J Med Chem. 2019;62(9):4411-4425.
Crossref - Venter H, Mowla R, Ohene-Agyei T, Ma S. RND-type drug efflux pumps from Gram-negative bacteria : molecular mechanism and inhibition. Front Microbiol. 2015;6:377.
Crossref - Thakur V, Uniyal A, Tiwari V. A comprehensive review on pharmacology of efflux pumps and their inhibitors in antibiotic resistance. Eur J Pharmacol. 2021;903:174151.
Crossref - Gholizadeh P, Kose S, Dao S, et al. How CRISPR-Cas system could be used to combat antimicrobial resistance. Infect Drug Resist. 2020;13:1111-1121.
Crossref - Dsouza N, Murthy NS, Aras RY. Projection of Cancer Incident Cases for India -Till 2026. Asian Pac J Cancer Prev. 2013;14:4379-4386.
Crossref - Garrett SC. Pruning and Tending Immune Memories: Spacer Dynamics in the CRISPR Array. Front Microbiol. 2021;12:664299.
Crossref - Gholizadeh P, Kose S, Dao S, et al. How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance. Infect Drug Resist. 2020;13:1111-1121.
Crossref - Pham TN, Loupias P, Dassonville-Klimpt A, Sonnet P. Drug delivery systems designed to overcome antimicrobial resistance. Med Res Rev. 2019;39(6):2343-2396.
Crossref - Lahiri D, Nag M, Dey A, Sarkar T, Pati S, Ray RR. Nanoparticles based antibacterial vaccines: Novel strategy to combat antimicrobial resistance. Process Biochem. 2022;119:82-89.
Crossref - Sivam V, Rangasamy A, Dara PK. Nanoparticle Approach to Control AMR. In: Handbook on Antimicrobial Resistance. Springer Nature Singapore. 2023:925-946.
Crossref - Vanamala K, Tatiparti K, Bhise K, et al. Novel approaches for the treatment of methicillin-resistant Staphylococcus aureus: Using nanoparticles to overcome multidrug resistance. Drug Discov Today. 2021;26(1):31-43.
Crossref - Wan F, Draz MS, Gu M, Yu W, Ruan Z, Luo Q. Novel Strategy to Combat Antibiotic Resistance: A Sight into the Combination of CRISPR/Cas9 and Nanoparticles. Pharmaceutics. 2021;13(3):352.
Crossref - Khambhati K, Bhattacharjee G, Gohil N, et al. Phage engineering and phage-assisted <scp>CRISPR-Cas</scp> delivery to combat multidrug-resistant pathogens. Bioeng Transl Med. 2023;8(2):10381.
Crossref - Zohra T, Numan M, Ikram A, et al. Cracking the Challenge of Antimicrobial Drug Resistance with CRISPR/Cas9, Nanotechnology and Other Strategies in ESKAPE Pathogens. Microorganisms. 2021;9(5):954.
Crossref - Mora-Ochomogo M, Lohans CT. β-Lactam antibiotic targets and resistance mechanisms: from covalent inhibitors to substrates. RSC Med Chem. 2021;12(10):1623-1639.
Crossref - Myers AG, Clark RB. Discovery of Macrolide Antibiotics Effective against Multi-Drug Resistant Gram-Negative Pathogens. Acc Chem Res. 2021;54(7):1635-1645.
Crossref - Stogios PJ, Savchenko A. Molecular mechanisms of vancomycin resistance. Protein Sci. 2020;29(3):654-669.
Crossref - Rusu A, Buta EL. The Development of Third-Generation Tetracycline Antibiotics and New Perspectives. Pharmaceutics. 2021;13(12):2085.
Crossref - Wang N, Luo J, Deng F, Huang Y, Zhou H. Antibiotic Combination Therapy: A Strategy to Overcome Bacterial Resistance to Aminoglycoside Antibiotics. Front Pharmacol. 2022;13:839808.
Crossref - Brenciani A, Morroni G, Schwarz S, Giovanetti E. Oxazolidinones: mechanisms of resistance and mobile genetic elements involved. J Antimicrob Chemother. 2022;77(10):2596-2621.
Crossref - Lungu IA, Moldovan OL, Biris V, Rusu A. Fluoroquinolones Hybrid Molecules as Promising Antibacterial Agents in the Fight against Antibacterial Resistance. Pharmaceutics. 2022;14(8):1749.
Crossref - Van Bambeke F, Balzi E, Tulkens PM. Antibiotic efflux pumps. Biochem Pharmacol. 2000;60(4):457-470.
Crossref - Lowrence RC, Subramaniapillai SG, Ulaganathan V, Nagarajan S. Tackling drug resistance with efflux pump inhibitors: from bacteria to cancerous cells. Crit Rev Microbiol. 2019;45(3):334-353.
Crossref - Huang L, Wu C, Gao H, et al. Bacterial Multidrug Efflux Pumps at the Frontline of Antimicrobial Resistance: An Overview. Antibiotics. 2022;11(4):520.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.